Using BioBERT and Qdrant to Power Semantic Search on Medical Q&A data
Navigating Complex Medical Datasets: Integrating BioBERT’s NLP with Qdrant’s Vector Database for Enhanced Semantic Accuracy
Photo by National Cancer Institute on Unsplash
In this tutorial, we’re diving into the fascinating world of powering semantic search using BioBERT and Qdrant with a Medical Question Answering Dataset from HuggingFace. We’ll unravel the complexities and intricacies of semantic search, a process that goes beyond mere keyword matching to understand the deeper meaning and context of queries.
Our journey will also explore the functionalities of Qdrant, a vector similarity search engine, in handling and extracting nuanced information from a rich medical dataset. BioBERT is a BERT-based language model specially designed for text-mining tasks in the healthcare(medicine) domain. The healthcare sector is in growing need of accurate and contextually relevant information retrieval as healthcare-related data is being amassed digitally.
But before we begin this tutorial, let’s first refresh our understanding of the key concepts it encompasses.
Introduction to Semantic Search
Semantic search is a search process employed by a search engine to yield relevant results through content and contextual mapping, instead of exact literal matches. The Wikipedia definition is as follows:
Semantic search denotes search with meaning, as distinguished from lexical search where the search engine looks for literal matches of the query words or variants of them, without understanding the overall meaning of the query.
Now, let me explain it in simpler terms. Semantic search is a bit like being a really good detective. It’s not just about looking for the exact words you typed into a search bar. Instead, it’s about understanding the meaning behind your words, almost like it’s trying to get into your head. Imagine you’re asking a friend about a book you can’t quite remember the name of. You describe it as “that book about the boy wizard with a lightning scar.” Your friend knows you’re talking about Harry Potter, even though you didn’t say it directly. That’s what semantic search does but on the internet.
This type of search looks at the context of your words, the relationships between them, and even the intent behind your query. It’s like it’s trying to understand the language the way humans do; not just as a list of keywords that needs to be matched.
Semantic search is important because it makes finding information easier and more natural. It’s like having a search engine that thinks more like a human and less like a robot, which is pretty cool. It helps you find what you’re looking for, even if you’re not sure how to ask for it perfectly. This is especially useful as the world’s information keeps growing. It’s like having a guide who not only knows the way but also understands why you’re asking the question and where you want to go.
About Qdrant
Semantic search is employed by search engines and databases to scour through the data efficiently. Search engines, especially those that handle loads of information, use semantic search to sort through mountains of data and find those needles in the haystack that are most relevant to your query.
One such search engine is Qdrant. It is a sophisticated open-source vector database engineered for scalability and efficiency in handling complex data searches. At its core, Qdrant utilizes vector embedding to represent data.
How does Qdrant power semantic searches
In simpler terms, Qdrant converts complex information like text, images, or even sound into numerical vectors — think of these as unique digital fingerprints. These vectors are not random; they are carefully calculated to represent the intrinsic properties of the data. This method allows Qdrant to compare and search through these vectors quickly and accurately.
The engine is optimized for high-performance similarity search, meaning it can swiftly sift through these digital fingerprints to find the closest matches to a query. This is crucial for applications requiring nuanced data understanding, like content recommendation systems or sophisticated search functionalities across large databases.
Once the vector embeddings are created from the raw data, it is stored in a vector database. The database then uses mathematical operations — such as Euclidean Distance, Cosine Similarity, Dot Product, etc. — to determine how similar the query is to the stored data. This results in highly accurate and context-aware search results.
Problem statement
The project aims to develop a semantic search engine to accurately retrieve medical information from a question-answering dataset. This search engine is intended to assist users in finding the most relevant medical answers to their queries, enhancing information accessibility and accuracy in the medical domain.
The approach involves vectorizing medical questions, answers, and contextual information using BioBERT, a language model pre-trained on biomedical texts. These vectors represent the semantic content of the text, enabling a similarity-based search. The vectorized data is indexed using Qdrant, a vector search engine, which allows for efficient similarity searches. The search functionality utilizes the vector representations to find the closest matches to a given query in the dataset. This usecase is part of the healthcare domain and therefore needs more precision. Qdrant helps ensure that when you’re looking for specific medical information, you’re getting the most accurate and relevant answers.
This method is chosen for its ability to capture the complex semantics of medical language and provide contextually relevant search results, going beyond keyword matching to understand the deeper meaning of medical queries.
Implementation of the solution
This approach is structured to harness the advanced NLP capabilities of BioBERT for semantic understanding and leverage Qdrant’s efficient search mechanism for retrieving contextually relevant medical information.
The following are the steps to the solution:
Loading the Dataset: Acquire and load the medical question-answering dataset from huggingface.
Preprocessing Textual Data: Normalize the data and apply lemmatization while preserving named entities, crucial for maintaining medical terminologies. This process ensures the integrity and specificity of medical information.
Vectorization Using BioBERT: Utilize the BioBERT model, specifically trained on biomedical literature, to convert text into semantic vectors. This model is chosen for its proficiency in understanding complex medical contexts.
Setting Up Qdrant Cloud: Create an account and a cluster on Qdrant Cloud, and set up QdrantClient for interaction.
Uploading Data: Create a collection on Qdrant Cloud. Index and upload vectors formed from a combination of context, question, and answer. This comprehensive vectorization captures the full scope of the information.
Implementing Search Functionality: Vectorize input queries using the same model and search in Qdrant, followed by result handling.
Testing the Search Functionality: Conduct tests with known and novel queries to evaluate the system’s effectiveness and ability to generalize.
Now, let's jump to the code.
Code
I have provided the code below for each section of the implementation with an explanation. If you just want to check the entire code, here is the link to github.
Pre-requisites
Install the requirements
Run the following code to install all the required libraries and the dataset
pip install qdrant-client
pip install transformers
pip install spacy
pip install https://huggingface.co/spacy/en_core_web_sm/resolve/main/en_core_web_sm-any-py3-none-any.whl
pip install torch
pip install datasets
pip install torch torchvision torchaudio
Import all the required libraries
from datasets import load_dataset
import spacy
import re
import pandas as pd
from transformers import AutoTokenizer, AutoModel
import torch
import numpy as np
from qdrant_client import QdrantClient, models
import numpy as np
from qdrant_client import models
Load dataset
The dataset used is loaded from huggingface. This dataset has questions and answers related to 8 topics in the medical field. Each question and answer also has a context column that provides a detailed description of the medical issue pertaining the Q&A pair. Check this huggingface link for a more thorough dataset description.
# The dataset URL
dataset_url = "GonzaloValdenebro/MedicalQuestionAnsweringDataset"
# Load the dataset
dataset = load_dataset(dataset_url)
#convert the dataset to pandas dataframe
df = dataset["train"].to_pandas()
Preprocessing the textual data
The dataset is processed in this step. Firstly, we will normalize the textual data which includes removing any lowercase and unnecessary punctuation. Then, lemmatization is applied while preserving named entities, crucial for maintaining medical terminologies. This process ensures the integrity of medical information.
nlp = spacy.load("en_core_web_sm")
def preprocess_text(text):
# Normalization: Lowercase and remove unnecessary punctuation
text = text.lower()
text = re.sub(r'[^\w\s]', '', text)
# Tokenization, Lemmatization and Named Entity Recognition with spaCy
doc = nlp(text)
processed_tokens = []
for token in doc:
if token.is_stop or token.is_punct:
continue
elif token.ent_type_: # Check if the token is a named entity
processed_tokens.append(token.text) # Preserve named entities as they are
else:
processed_tokens.append(token.lemma_) # Lemmatize non-entity tokens
# Re-join tokens
return ' '.join(processed_tokens)
# Assuming df is your DataFrame
df['Processed_Question'] = df['Question'].apply(preprocess_text)
df['Processed_Context'] = df['Context'].apply(preprocess_text)
df['Processed_Answer'] = df['Answer'].apply(preprocess_text)
Vectorization Using BioBERT
Loading BioBERT tokenizer and model
In this step, we will load the BioBERT model and the tokenizer which will help us to vectorize the pre-processed textual data. We are using the BioBERT model to vectorize the data because it is specifically trained on biomedical literature, to convert text into semantic vectors. This model is chosen for its proficiency in understanding complex medical contexts.
tokenizer = AutoTokenizer.from_pretrained("dmis-lab/biobert-base-cased-v1.1")
model = AutoModel.from_pretrained("dmis-lab/biobert-base-cased-v1.1")
Vectorize data
In this step, we will vectorize the dataset using the bioBERT tokenizer. Vectorization is a critical step in natural language processing, especially for tasks like semantic search. It is essential for semantic search because it transforms text into a mathematical representation that algorithms can process. By representing text as vectors, mathematical methods like cosine similarity can evaluate how semantically similar they are. This method allows us to effectively capture and compare the nuances of meaning within the text.
def vectorize_text(text, tokenizer, model):
# Tokenize and encode the text
inputs = tokenizer(text, return_tensors="pt", padding=True, truncation=True, max_length=512)
# Move tensors to the same device as the model
inputs = {k: v.to(model.device) for k, v in inputs.items()}
# Get the output from the model
with torch.no_grad():
outputs = model(**inputs)
# Use the mean of the last hidden states as the vector representation
return outputs.last_hidden_state.mean(dim=1).squeeze().cpu().numpy()
df['Vectorized_Question'] = df['Processed_Question'].apply(lambda x: vectorize_text(x, tokenizer, model))
df['Vectorized_Context'] = df['Processed_Context'].apply(lambda x: vectorize_text(x, tokenizer, model))
df['Vectorized_Answer'] = df['Processed_Answer'].apply(lambda x: vectorize_text(x, tokenizer, model))
Setting Up Qdrant Cloud
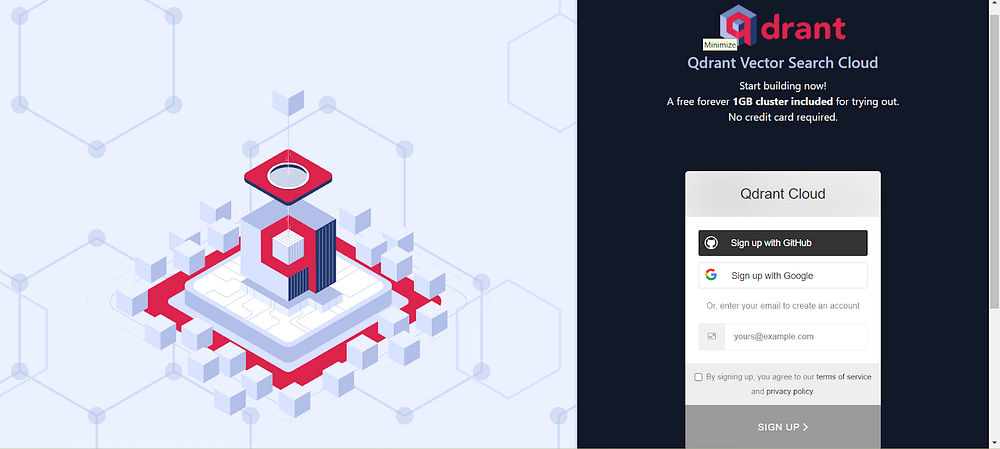
Create an account on Qdrant Cloud
- Sign up for an account on Qdrant using this link.
Image by Author
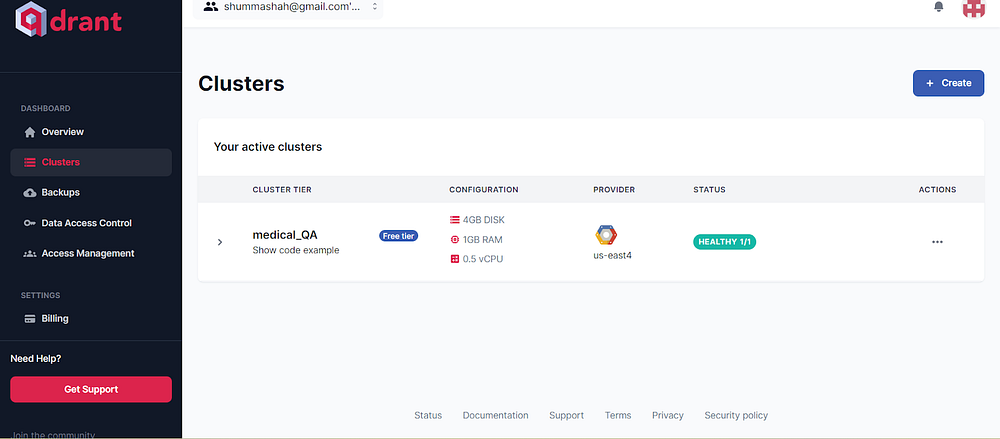
Create a new cluster. Under Set a Cluster Up enter a Cluster name. I have named my cluster “medical_QA”.
Click Create Free Tier and then Continue.
Under Get an API Key, select the cluster and click Get API Key.
Save the API key, as you won’t be able to request it again. Click Continue.
Save the code snippet provided to access your cluster. Click Complete to finish setup.
The created cluster will look like this
Image by Author
Set Up QdrantClient
Use the code snippet you received in the previous step to access your cluster.
qdrant_client = QdrantClient(
url="https://1259bacd-03fe-4c21-992e-383f0e51fd47.us-east4-0.gcp.cloud.qdrant.io:6333",
api_key="iKfB4nUdjCSrvuQZIrfQcMBdacJHVWybK1NGaJzCiKCx42jNbnCc7w"
)
Uploading Data
Create a collection on Qdrant cloud
All data in Qdrant is organized in collections. So we will create a collection using recreate_collection. It is used when experimenting as it first deletes a collection with the same name. Vector_size parameter defines the size of the vectors for a collection. The distance parameter allows us to calculate the distance between two points using different methods like Euclidean Distance, Cosine Similarity, etc. Here we used Cosine similarity to measure the distance. This metric measures the cosine of the angle between two non-zero vectors in a multi-dimensional space.
# Define the collection name and vector size (depends on BioBERT model, e.g., 768 for base models)
collection_name = "semantic_search_medical_qa"
vector_size = 768 # Adjust based on your BioBERT model
# Create a collection
qdrant_client.recreate_collection(
collection_name=collection_name,
vectors_config = models.VectorParams(
size=vector_size, # Vector size is defined by used model
distance=models.Distance.COSINE,
)
)
Create a vectorized data list
Before uploading the data to the cloud, we need to create the vectorized data list. The “vector” field in this list is formed using a vectorized combination of context, question, and answer. The other fields are the raw textual data of the question, answer, and context.
Instead of just vectorizing the answer, I vectorized all the fields in order to capture the full scope of the information related to the question. This will help to establish a more comprehensive link between the query and the answer. The ‘vector’ field within the vectorized_data list serves as the numerical representation of the text data, combining the semantic essence of the question, context, and answer. In Qdrant, this enables the database to perform similarity searches. When a query is vectorized and compared against these stored vectors, Qdrant can then efficiently retrieve the most semantically relevant entries by calculating the proximity between the query vector and document vectors, typically using cosine similarity as the metric. Since I have vectorized all the necessary information, the retrieved data after proximity calculation will be more accurate.
vectorized_data = [
{
'id': row['id'],
'vector': np.concatenate([
np.fromstring(row['Vectorized_Question'].replace('\n', '').replace('[', '').replace(']', ''), sep=' '),
np.fromstring(row['Vectorized_Context'].replace('\n', '').replace('[', '').replace(']', ''), sep=' '),
np.fromstring(row['Vectorized_Answer'].replace('\n', '').replace('[', '').replace(']', ''), sep=' ')
]),
'question': row['Question'],
'context': row['Context'],
'answer': row['Answer']
}
for _, row in df.iterrows()
]
Upload vectors to the cloud
We are uploading the vectors to the cloud in batches. Given the large size of the dataset, batch processing reduces the risk of overloading the network and server capacity, and ensures no error is received. I have selected 50 to be the batch size, you can reduce it further incase you receive any network error.
For this demonstration, I have uploaded only 60% of the dataset to showcase the functionality while conserving time. You can use the same method to upload the entire dataset, ensuring completeness of the data in Qdrant without compromising the upload process. Make sure to replace the subset_size with len(vectorized_data) in the range function and print statement while uploading the dataset to ensure the upload of 100% of the dataset.
collection_name = "semantic_search_medical_qa" # mention the collection name
def upload_batch(batch):
# Upload the batch to Qdrant
qdrant_client.upload_records(
collection_name=collection_name,
records=[
models.Record(
id=int(data_point["id"]),
vector=data_point["vector"].tolist(), # Convert numpy array to list
payload={
"question": data_point["question"],
"context": data_point["context"],
"answer": data_point["answer"]
}
)
for data_point in batch
],
)
# Batch size
batch_size = 50
# Calculate 60% of the dataset size
subset_size = int(len(vectorized_data) * 0.6)
# Upload the dataset
for i in range(0, subset_size, batch_size):
batch = vectorized_data[i:i + batch_size]
upload_batch(batch)
print(f"Total no of batches {subset_size/batch_size}")
print(f"Uploaded batch {i // batch_size}")
Implement Search Functionality
Search the database
We will first create a function that vectorizes all the input queries just like we did for the raw data. Now, we will set up the function that searches the qdrant database.
def vectorize_query(query, tokenizer, model):
inputs = tokenizer(query, return_tensors="pt", padding=True, truncation=True, max_length=512)
with torch.no_grad():
outputs = model(**inputs)
return outputs.last_hidden_state.mean(dim=1).squeeze().cpu().numpy()
def search_in_qdrant(query, tokenizer, model, top_k=10):
query_vector = vectorize_query(query, tokenizer, model)
# Search in Qdrant
hits = qdrant_client.search(
collection_name="semantic_search_medical_qa",
query_vector=query_vector.tolist(),
limit=top_k,
)
return hits
Result Handling
Now, we will create a function that displays the search results based on the cutoff score. The score in Qdrant’s search results generally signifies the similarity between the query vector and the vectors in the database. Since we used cosine similarity, the score represents how close the query is to each result in terms of the cosine of the angle between their vectors. A higher score indicates greater similarity.
def display_search_results(test_query, tokenizer, model, cutoff_score):
results = search_in_qdrant(test_query, tokenizer, model)
for result in results:
if result.score >= cutoff_score:
print("Answer:", result.payload["answer"])
print("Context:", result.payload["context"])
print("Score:", result.score)
print("-----------")
Results
Testing the Search Functionality
Now we will test this using three different queries and check how it performs.
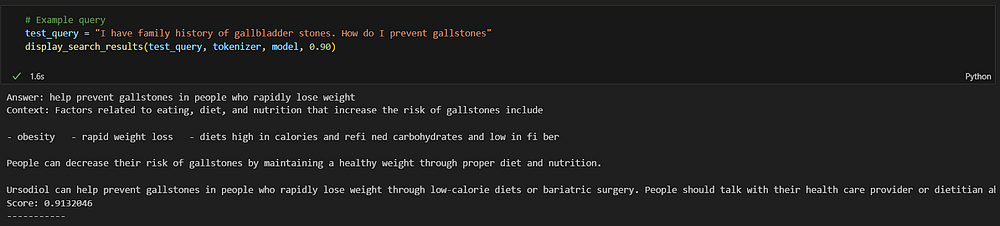
Example Query 1
test_query = "I have a family history of gallbladder stones. How do I prevent gallstones"
display_search_results(test_query, tokenizer, model, 0.90)
The query I have asked is not part of the original database but still we got the relevant answer. I am filtering out the responses based on a 90% similarity score.
Image by Author
Example Query 2
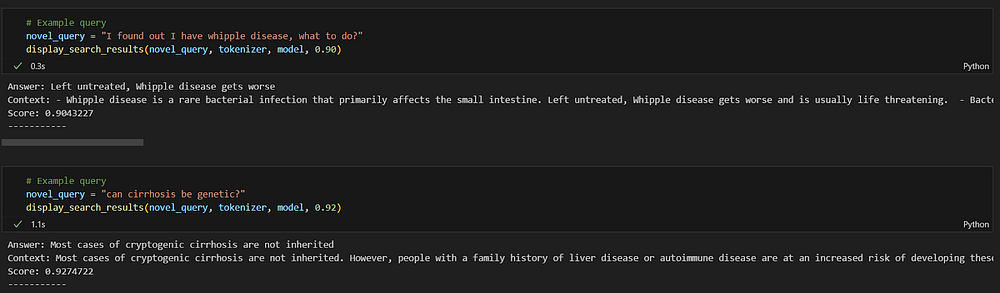
novel_query = "I found out I have whipple disease, what to do?"
display_search_results(novel_query, tokenizer, model, 0.90)
In this scenario, I asked a question as a patient who wants to know the treatment of Whipple disease. In order to test the semantic search critically, I avoided the use of keywords like “treatment” and instead used informal spoken language like “what to do”.
Despite this, the response is the most relevant and correct amongst the database.
Image by Author
Example Query 3
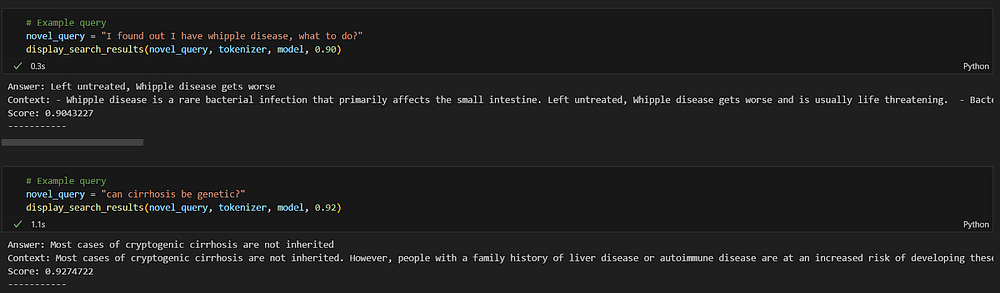
novel_query = "can cirrhosis be genetic?"
display_search_results(novel_query, tokenizer, model, 0.92)
In this query, I used the word genetic but still the search correctly provided a response where “family history” is mentioned. It correctly identified the contextual link between the words genetic and family history. I filtered the response based on 92% similarity and got the most relevant answer.
Image by Author
Conclusion
The semantic search engine, utilizing BioBERT for vectorization, has demonstrated an impressive ability to interpret and retrieve medically relevant answers. The process involved meticulous steps, from data preprocessing and vectorization to setting up Qdrant Cloud and implementing robust search functionality. The outcome is a search engine that not only understands the nuances of medical queries but also provides relevant and accurate answers.
The testing phase with varied queries shows that the system effectively understands and responds to the context of inquiries. For instance, the query about gallbladder stones prevention returned a highly relevant answer without direct reference in the database. Similarly, a query about Whipple disease was met with an accurate response, despite the use of informal language. Furthermore, the system’s adeptness at contextually linking “genetic” to “family history” in the cirrhosis query underscores its capability to discern semantic connections. These results highlight the engine’s potential as a robust tool for providing accurate medical information which further goes on to showcase its understanding of complex medical terminologies and patient inquiries.
As we conclude, it’s clear that the marriage of advanced NLP models and semantic search engines like Qdrant opens new horizons for information retrieval, promising more accurate, efficient, and context-aware solutions for complex search scenarios that have high stakes like healthcare.
Thank you for reading this article! If you enjoyed the content and would like to stay in the loop on future explorations into technology, AI, and beyond, please follow me on LinkedIn.
On my LinkedIn profile, I regularly delve into topics lying at the intersection of AI, technology, data science, personal development, and philosophy.
I’d love to connect and continue the conversation with you there.